


TransProg is a set of facilities (multiplatform and writen in C language) for a programmer to harness the full potential of modern general purpose computers.
It was designed with individual-based simulations in mind, and consequently makes use of multiple cores and processors, as well as graphical processing units for both rendering and computing.
It is dedicated to interactive simulations of highly dynamic systems where entities can move, change, appear, disappear and interact with each other and the user at any time.
Experiments with various computers, operating systems and compilers yield very satisfying results in terms of performance and scalability relating to the number of CPUs used.
A dynamic load-balancing mechanism, taking into account the
applicative affinity between entities ensures good data locality
within cache memories.



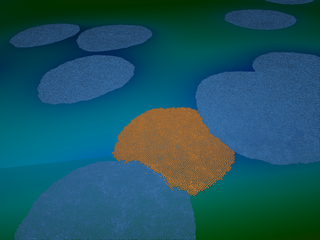
Example: interactive simulation of millions of bacteria.
The substrate is 1.783 cm wide and 1 cm thick.
Made of 1 656 837 spatial cells organised as a 357x357 regular horizontal
grid and 13 planes with a geometrical progression, it is responsible for
glucose and lactate diffusion and pH computation.
Bacteria are from two types, with different sets of parameters, but each
individual consumes glucose, to grow and duplicate itself, and produces
lactate which in turn limits growth and duplication.
Moreover, each individual interacts with its nearest neighbours (around
twenty) according to attraction/repulsion rules.
The following pictures and video show 16 million bacteria but this
simulation reaches memory exhaustion beyond 262 million bacteria on a
dual Intel Xeon E5-2697 v2 computer with 16GB RAM.

 (20 MB video)
(20 MB video)
Layers of bacteria :
7 MB video


The source code (TransProg_2016m07d04.tgz) is released under the GNU LGPL license.
The following article describes the design of this library:
The various works described in these articles make use of this library: